
点击右上角![]() 微信好友
微信好友
 朋友圈
朋友圈

请使用浏览器分享功能进行分享


棉花是我国重要的经济作物和战略物资之一,新疆地区占有全国70%以上的植棉面积,产出约90%的皮棉,在棉花生产上具有举足轻重的地位。
得益于先进的制种工艺和高效的栽培耕作管理手段,新疆的棉花生产从大面积精量播种,化学调控到最后收获已经基本实现全面机械化,大大降低了棉农的劳动强度,真正实现了棉花科技工作者努力追求的快乐植棉的目标。

借助我国自主研发的北斗定位系统,可使播种机自动在棉田行进并进行单粒精量播种


成体系的种植模式使得大型化控机械和无人机可进入棉田实现大面积化学调控

到棉花成熟之际,一望无际的棉田是新疆植棉区的一道风景线

大型采棉机集中采收之后就可进行收纳运输
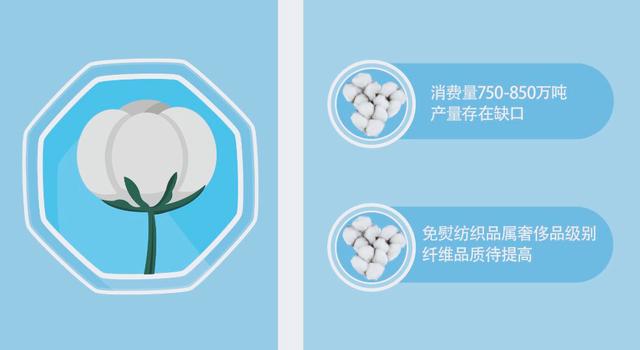
但新疆的棉花依然存在一定的问题,比如产量缺口大,品质相对低等,许多棉花科研工作者为了解决棉花生产问题,投入了大量的心血,并取得了重要的成果,我们一起来学习吧!

王茂军教授从事棉花基因组学研究,分析棉花纤维产生的进化机理
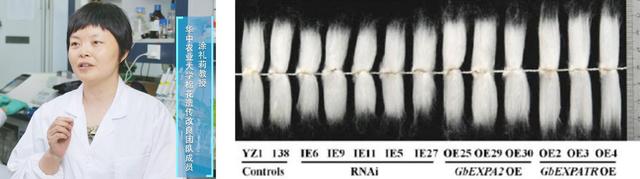
涂礼莉教授关注纤维发育过程,尝试利用长纤维海岛棉基因改良陆地棉
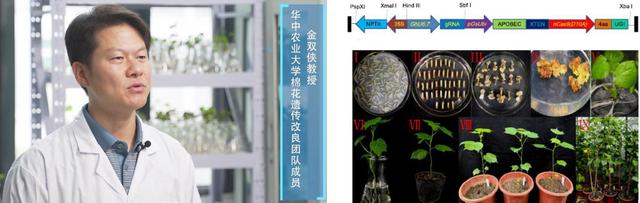
金双侠教授建立了棉花高效体遗传转化系统并开发了系列基因编辑工具,为深入研究棉花基因功能提供了重要手段,为加速棉花分子育种提供了有力工具
华中农业大学棉花遗传改良团队的三位老师分别从基因组学、分子生物学和生物技术三个方面出发,为大家讲解了棉花生产上的问题,并提出了相对应的解决方案。

棉花的作用远不止如此,种子丰富的营养元素和特殊的种质资源使得棉花有望成为集纤维、食品、观赏价值于一身的明星作物。越来越先进的人工智能和生物技术,正在为棉花研究添砖加瓦,中国棉花未来可期!
参考论文链接
(1)Maojun Wang, Lili Tu, Min Lin, et al. Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication, Nature Genetics. 49 (2017) 579–587 (https://www.nature.com/articles/ng.3807)
(2)Maojun Wang, Lili Tu, Daojun Yuan, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense, Nature Genetics. 51 (2019) 224–229. (https://www.nature.com/articles/s41588-018-0282-x)
(3)Maojun Wang, Pengcheng Wang, Lili Tu, et al. Multi-omics maps of cotton fibre reveal epigenetic basis for staged single-cell differentiation. Nucleic Acids Research. 44 (2016) 4067–4079, https://doi.org/10.1093/nar/gkw238. (https://academic.oup.com/nar/article/44/9/4067/2462395?searchresult=1)
(4)Yang Li, Lili Tu, Filomena A Pettolino, et al. GbEXPATR, a species-specific expansin, enhances cotton fibre elongation through cell wall restructuring.Plant Biotechnology Journal,14(2016)951-963. (https://onlinelibrary.wiley.com/doi/10.1111/pbi.12450)
(5)Fenglin Deng, Lili Tu, Jiafu Tan, et al.GbPDF1 Is Involved in Cotton Fiber Initiation via the Core cis-Element HDZIP2ATATHB2. Plant Physiology, 158 (2012) 890–904, https://doi.org/10.1104/pp.111.186742. (https://academic.oup.com/plphys/article/158/2/890/6109269)
(6)Qiongqiong Wang,Muna Alariqi,Fuqiu Wang, et al.The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnology Journal.18 (2020) 2436-2443. (https://onlinelibrary.wiley.com/doi/10.1111/pbi.13417)
(7)Lei Qin,Jianying Li,Qiongqiong Wang, et al. High-efficient and precise base editing of CG to TA in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnology Journal. 18 (2019) 45-56. (https://onlinelibrary.wiley.com/doi/10.1111/pbi.13168)
(8)Bo Li,Hangping Rui,Yajun Li, et al. Robust CRISPR/Cpf1 (Cas12a)-mediated genome editing in allotetraploid cotton (Gossypium hirsutum). Plant Biotechnology Journal. 17 (2019) 1862-1864. (https://onlinelibrary.wiley.com/doi/10.1111/pbi.13147)
(9)Pengcheng Wang, Jun Zhang, Lin Sun, Yizan Ma, Jiao Xu, Sijia Liang, Jinwu Deng, Jiafu Tan, Qinghua Zhang, Lili Tu, Henry Daniell, Shuangxia Jin, Xianlong Zhang. High efficient multi-sites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system, Plant Biotechnology Journal, 2018, 16, 137-150(https://onlinelibrary.wiley.com/doi/full/10.1111/pbi.1275)
(10)Jianying Li, Maojun Wang, Yajun Li, Qinghua Zhang, Keith Lindsey, Henry Daniell, Shuangxia Jin*, Xianlong Zhang. Multi-omics analyses reveal epigenomics basis for cotton somatic embryogenesis through successive regeneration acclimation (SRA) process, Plant Biotechnology Journal, 2019, 17, 435–450(https://onlinelibrary.wiley.com/doi/10.1111/pbi.12988)
出品:科普中国 中国作物学会 华中农业大学 光明网
编导:马益赞 徐琴
拍摄:任文武 龚时峰 戴壮
剪辑:龚时峰 戴壮
后期制作:任文武 龚时峰 戴壮
监制:张献龙 程维红
特别鸣谢:新疆农科院经济作物研究所 孔杰 郑巨云
来源: 中国作物学会
